Genetika-guppy.my1.ru
Меню сайта
Разделы дневника
| Генетика гуппи [9] |
| Селекция гуппи [9] |
| Генетика меченосца [0] |
| Селекция меченосца [0] |
| Гентика пецилии [0] |
| Селекция пецилии [0] |
| Генетика моллинезии [0] |
| Селекция моллинезии [0] |
| Генетика других живородящих рыб [0] |
| Селекция других живородящих рыб [0] |
Календарь
| « Январь 2009 » | ||||||
| Пн | Вт | Ср | Чт | Пт | Сб | Вс |
| 1 | 2 | 3 | 4 | |||
| 5 | 6 | 7 | 8 | 9 | 10 | 11 |
| 12 | 13 | 14 | 15 | 16 | 17 | 18 |
| 19 | 20 | 21 | 22 | 23 | 24 | 25 |
| 26 | 27 | 28 | 29 | 30 | 31 | |
Поиск
Друзья сайта
Мини-чат
Наш опрос
Статистика
Онлайн всего: 1
Гостей: 1
Пользователей: 0

БлогиБлоги
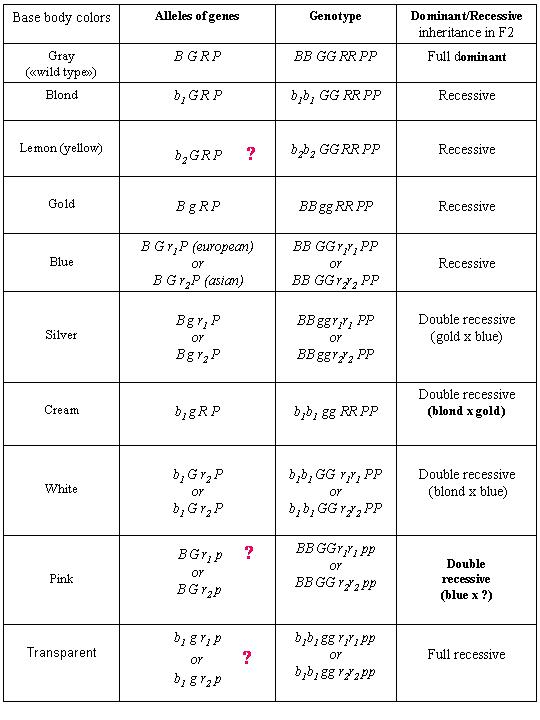
Главная » 2009 Январь 24 » Table "Base body colors guppy's genes"
Table "Base body colors guppy's genes" | 21:43 |
Table "Base body color guppy's genes"
 | |
| Категория: Генетика гуппи | Просмотров: 3986 | Добавил: genetika-guppy | |




